Analyze fits
Check if run correctly
In the output, you should have got the following directory structure:
master_outdir/ # Directory specified with "master_outdir" parameter in params.py
config_prefix1/ # named after config_prefix specifications in fitmap_args in params.py
map1.mrc/ # named after the filenames of the EM maps used
map1.mrc # symbolic link to the reference map
pdb_file_name1.pdb/ # named after the pdb file names used for fitting
solutions.csv # the list of solutions and their scores
solutions_pvalues.csv # the list of solutions and their scores including pvalues THIS IS THE FILE NEEDED FOR THE NEXT STEP
log_err.txt # standard error log
log_out.txt # standeard output log
run.sh # sbatch script used for running the job
ori_pdb.pdb # symbolic link to the original query file
map1.mrc # symbolic link to the reference map
Rplots.pdf # some statistics from the pvalue calculation
pdb_file_name2.pdb/
pdb_file_name3.pdb/
config.txt # A config file for fitting, saved FYI.
map2.mrc/
config_prefix2/
config_prefix3/
Check if you obtained these files, in particular the solutions_pvalues.csv file.
Note that ori_pdb.pdb and map1.mrc files are symbolic links to the original files. If for any reason those links are broken or do not exist, you can re-generate them by running the fit.py script with ``–update_links `` option:
fit.py --update_links efitter_params.py
Generating fitted PDBs
Although not necessary for the subsequent modelling, you can generate top fits as PDB files visualization.
You may for example see that for some structures you obtain significant p-values in solutions_pvalues.csv file,
and upon visual inspection, decide that you want to restrict the fits to these significant fits using max_positions parameter when Adding precomputed fitting libraries to JSON.
Method 1
Enter the results directory for the given map:
cd master_outdir/config_prefix1/map1.mrc/
Generate PDBs for multiple structures and/or maps into a single directory:
cd fit_cam_inside0.3_fa_10000 genPDBs_many.py [options] outdir <solutions_list>
Example case-scenario following:
Generate fits for all maps:
Enter the “parameters-set” fit directory like search100000_metric_cam_inside0.3_radius500/ and run:
genPDBs_many.py -n5 top5 */*/solutions.csv
This will generate a directory top5 with subdirectories for each map, and top 5 fits for each map.
Generate fits for a specific map:
Enter the directory for specific map like search100000_metric_cam_inside0.3_radius500/P_negstain_01.mrc and run:
genPDBs_many.py -n5 top5 */solutions.csv
This will generate a directory top5 with top 5 fits for each map
Method 2
A bash command like the following will iterate through all output directories and generate 10 fits there:
for f in `ls fits_chimera/fit_cam_inside0.3_Big/nr_8_norm_m3i_filt.no_membrane.mrc/* | grep ":" | perl -p -e "s/\://"`; do cd $f; genPDBs.py -n 10 solutions.csv ori_pdb.pdb cd /g/kosinski/kosinski/NPC/fitting/Chlamy done
Method 3 (“Manually”)
Enter the directory for the given run:
cd master_outdir/config_prefix1/map1.mrc/pdb_file_name1.pdb/
Generate the PDBs for a specified number of top fits:
genPDBs.py -n 5 solutions.csv <path to the pdb files>/pdb_file_name1.pdb
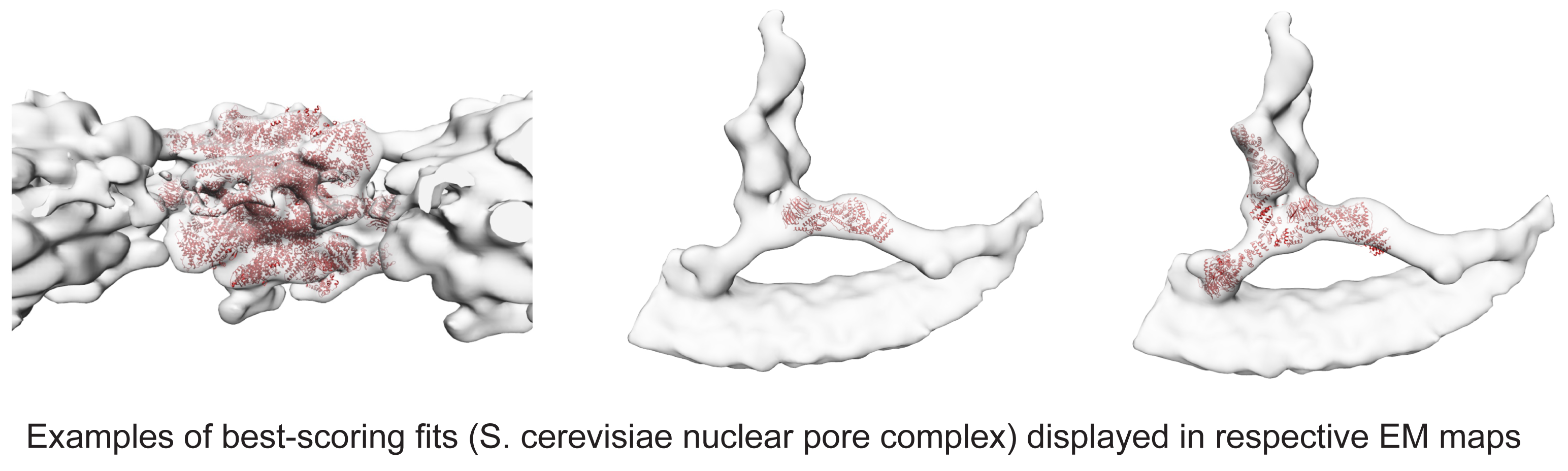
Visualize the pdbs with your map in Chimera (example figure following)